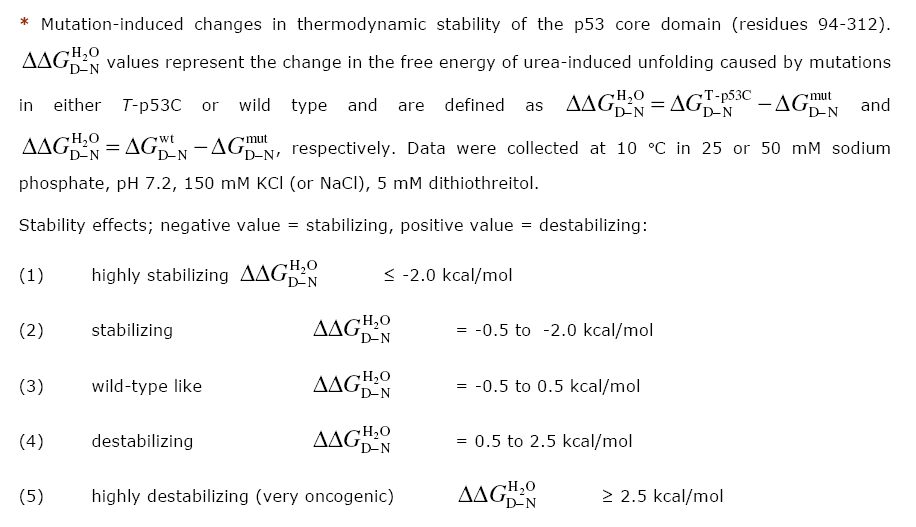
| Mutant | ddG* | dG Ref* | Stability* | Reference Stability | PDB structure | Reference structure | Crystallography |
|---|---|---|---|---|---|---|---|
| Q104H | -0.24 | WT | wild-type like | Nikolova PV, 1998 | |||
| Q104P | -0.11 | WT | wild-type like | Nikolova PV, 1998 | |||
| T123A | 0.13 | WT | wild-type like | Nikolova PV, 2000 |
Thr-123 is located at the end of the flexible L1
loop that makes contacts with the DNA major groove via Lys-120. The Thr-123 side chain is
solvent exposed.
Its hydroxyl group makes no direct but water-mediated intramolecular contacts. Apart from
the truncated side
chain (threonine to alanine), no significant T123A-induced structural changes are observed
in the DNA-free
crystal structure of the heptamutant T123A/M133L/H168R/
V203A/N239Y/R249S/N268D (PDB 2BIQ) when compared with the structure of the corresponding hexamutant without T123A (PDB 2BIP). |
||
| A129D | 0.7 | WT | destabilizing | Nikolova PV, 1998 | |||
| A129E | 0.38 | WT | wild-type like | Nikolova PV, 1998 | |||
| A129S | 0.19 | WT | wild-type like | Nikolova PV, 1998 | |||
| M133L | -0.3 | WT | wild-type like | Nikolova PV, 1998 |
Met-133 is located on β-strand S2’
of the loop-sheet-helix motif. Via its side-chain it contributes to the central hydrophobic
core of the protein. The M133L mutation induces only minor structural changes in the
immediate
environment of the mutation and leaves the overall structure of the hydrophobic core intact
(PDB 1UOL).
|
||
| F134L | 4.78 | WT | highly destabilizing | Bullock AN, 2000 | |||
| V143A | 3.7 | T | highly destabilizing | Joerger AC, 2006 | 2J1W | Joerger AC, 2006 |
Val-143 is located on β-strand S3.
Its side chain is part of the central hydrophobic core of the β-sandwich. The V143A
mutation
creates an internal cavity in the hydrophobic core of the β-sandwich without collapse
of the surrounding
structure. The mutant is highly destabilized as a result of lost hydrophobic interactions
due to the
two truncated methyl groups (PDB 2J1W).
|
| V143A | 3.5 | WT | highly destabilizing | Bullock AN, 2000 | |||
| L145Q | 2.98 | WT | highly destabilizing | Bullock AN, 2000 | |||
| D148E | 0.43 | WT | wild-type like | Nikolova PV, 1998 | |||
| D148S | -0.22 | WT | wild-type like | Nikolova PV, 1998 | |||
| T150P | 0.08 | WT | wild-type like | Nikolova PV, 1998 | |||
| P151S | 4.49 | WT | highly destabilizing | Bullock AN, 2000 | |||
| V157F | 3.88 | WT | highly destabilizing | Bullock AN, 2000 | |||
| Q165K | 1.27 | WT | destabilizing | Nikolova PV, 1998 | |||
| Q167E | 0.43 | WT | wild-type like | Nikolova PV, 1998 | |||
| H168R | 2.75 | WT | highly destabilizing | Nikolova PV, 2000 |
The highly destabilizing H168R mutation induces
substantial structural distortion around the mutation site in the L2 loop at the periphery
of the
DNA-binding surface. Several residues including the mutation site are disordered in the
crystal structure;
i.e. no defined conformation is observed for residues 166-170 and the side chain of Glu-171
(PDB 2BIN).
|
||
| H168R | 3.07 | T | highly destabilizing | Joerger AC, 2005 | 2BIN | Joerger AC, 2005 | |
| R174K | 0.22 | WT | wild-type like | Nikolova PV, 1998 | |||
| R175A | 0.73 | WT | destabilizing | Bullock AN, 2000 | |||
| R175H | 2.5 | T | highly destabilizing | Ang HC, 2006 | |||
| R175H | 3.52 | WT | highly destabilizing | Bullock AN, 2000 | |||
| C182S | -0.16 | WT | wild-type like | Nikolova PV, 1998 | |||
| I195T | 4.12 | WT | highly destabilizing | Bullock AN, 2000 | |||
| L201P | -0.35 | WT | wild-type like | Nikolova PV, 1998 | |||
| V203A | -0.49 | WT | stabilizing | Nikolova PV, 1998 |
Val-203 is located in the turn connecting the β-strands
S5 and S6. Mutation to alanine has no effect on the overall structure of the protein apart
from conformational changes of adjacent side chains (PDB 1UOL).
|
||
| L206S | 0.1 | WT | wild-type like | Nikolova PV, 1998 | |||
| Y220C | 4.2 | T | highly destabilizing | Joerger AC, 2006 | 2J1X | Kitayner M, 2006 |
The Y220C mutation is located at the far end of
the β-sandwich, at the start of the loop connecting β-strands S7 and S8. This
highly
destabilizing mutation creates an extended surface crevice but retains the structural
features of the
wild type in functionally important surface regions (PDB 2J1X).
|
| Y220C | 3.98 | WT | highly destabilizing | Bullock AN, 2000 | |||
| D228E | -0.05 | WT | wild-type like | Nikolova PV, 1998 | |||
| I232T | 3.19 | WT | highly destabilizing | Bullock AN, 2000 | |||
| Y236F | -0.27 | WT | wild-type like | Nikolova PV, 1998 | |||
| M237I | 3.18 | WT | highly destabilizing | Bullock AN, 2000 | |||
| N239Y | -1.49 | WT | stabilizing | Nikolova PV, 1998 | 2AC0 | Joerger AC, 2006 |
Asn-239 is located at the beginning of the L3 loop in the
immediate vicinity of the zinc-binding site. Its side chain is solvent exposed and makes
water-mediated
contacts with the DNA backbone upon binding of specific p53 response elements (e.g. PDB
2AC0). The N239Y
mutation stabilizes the protein and rigidifies the local structure without perturbing it. It
creates novel
hydrophobic packing interactions between the zinc-binding region and Leu-137 at the edge of
the
loop-sheet-helix motif (S2’/S3 loop) (PDB 1UOL).
|
| C242S | 3.07 | WT | highly destabilizing | Bullock AN, 2000 | |||
| G245S | 0.8 | T | destabilizing | Ang HC, 2006 | 2J1Y | Joerger AC, 2006 |
Gly-245 is located in the L3 loop, close to the
zinc-binding site. It adopts a main-chain conformation that is not favored in non-glycine
residues. Upon
G245S mutation, the serine side chain is accommodated by displacing a structural water
molecule that is
conserved in the wild type and other mutant structures. The mutation is accompanied by small
but significant
changes in the backbone conformation of the L3 loop. The structural changes affect residues
that form part
of the self-complementary core domain-core domain dimerization interface in the p53-DNA
complex. Presumably
G245S impairs DNA binding by reducing binding cooperativity (PDB 2J1Y).
|
| G245S | 1.21 | WT | destabilizing | Bullock AN, 2000 | |||
| G248Q | 1.87 | WT | highly destabilizing | Bullock AN, 2000 | |||
| R249S | 1.92 | WT | destabilizing | Bullock AN, 2000 |
The guanidinium group of Arg-249 makes crucial
interactions that stabilize the hairpin conformation of the L3 loop, which docks the core
domain to the
minor groove of DNA-response elements via Arg-248. It forms a salt bridge with Glu-171 and
hydrogen bonds
with the backbone oxygens of Gly-245 and Met-246. Upon mutation to serine, these
interactions are lost.
The L3 loop becomes highly flexible and undergoes a large conformational change, favoring a
non-native
conformation. Both DNA contacts (Arg-248) and residues that form the self-complementary core
domain-core
domain interface upon DNA binding (e.g. Met-243 and Gly-244) are affected. Most notable is a
switch of
methionines 243 and 246. Met-243 is located on the surface of the core domain in the wild
type.
In the R249S mutant, this region adopts a helical conformation, and the side chain of
Met-243 is
buried in the interior of the protein by displacing the side chain of Met-246 from this
buried position
(PDB 2BIO).
|
||
| R249S | 1.98 | T | destabilizing | Joerger AC, 2005 | 2BIO | Joerger AC, 2005 | |
| I255F | 3.29 | WT | highly destabilizing | Bullock AN, 2000 | |||
| S260P | 0.32 | WT | wild-type like | Nikolova PV, 1998 | |||
| N268D | -1.21 | WT | stabilizing | Nikolova PV, 2000 | Asn-268 is located in β-strand S10. The stabilizing N268D mutation results in an altered hydrogen bond pattern, linking the two sheets of the β-sandwich in an energetically more favorable way than in the wild type. In the mutant, the side chain of Asp-268 has flipped relative to the position of the asparagine in the wild type, and its carboxylate group is hydrogen-bonded to the main-chain amides of Leu-111 and Ser-269, thus stabilizing the protein (PDB 1UOL). | ||
| F270C | 4.54 | WT | highly destabilizing | Bullock AN, 2000 | |||
| F270L | 4.1 | T | highly destabilizing | Joerger AC, 2006 | 2J1Z | Joerger AC, 2006 |
Phe-270 is located on β-strand S10. Its side
chain is an integral part of the central hydrophobic core of the β-sandwich. The F270L
mutation
creates an internal cavity in this hydrophobic core without collapse of the surrounding
structure. The
mutant is highly destabilized as a result of the truncated hydrocarbon moiety (PDB 2J1Z).
|
| R273C | -0.4 | T | wild-type like | Joerger AC, 2006 | 2J20 | Joerger AC, 2006 |
R273C removes the essential DNA-contact residue Arg-273 that
contacts the phosphate backbones at the center of a DNA half-site. The conformation of
neighboring side
chains (e.g. Phe-134, Ser-240 or Asp-281) has not significantly changed in the mutant
crystal structure,
i.e. the overall architecture of the DNA-binding surface is preserved (PDB 2J20).
|
| R273H | 0.45 | WT | wild-type like | Bullock AN, 2000 |
R273H removes the essential DNA-contact residue
Arg-273 that contacts the phosphate backbone at the center of a DNA half-site. The
conformation of
neighbouring side chains (e.g. Phe-134, Ser-240 or Asp-281) has not significantly changed in
the mutant
crystal structure, i.e. the overall architecture of the DNA-binding surface is preserved
(PDB 2BIM).
|
||
| R273H | 0.09 | T | wild-type like | Joerger AC, 2005 | 2BIM | Joerger AC, 2005 | |
| R282W | 3.3 | WT | highly destabilizing | Bullock AN, 2000 |
In the wild-type protein, Arg-282 stabilizes the
loop-sheet-helix motif via a network of hydrogen bonds and hydrophobic interactions (e.g.
hydrogen
bonds with the hydroxyl group of Thr-125 and the backbone oxygen of Tyr-126, and a salt
bridge with Glu-286).
These interactions are lost in the R282W mutant, which is substantially destabilized as a
result. The
tryptophan side chain causes steric hindrance and perturbs the L1 loop, which includes the
DNA-contact
residue Lys-120. The overall architecture of the remainder of the DNA-binding region is
conserved,
consistent with the observation that the R282W mutant binds to several promoters at
sub-physiological
temperature at which the mutant is folded (PDB 2J21).
|
||
| R282W | 3 | T | highly destabilizing | Ang HC, 2006 | 2J21 | Joerger AC, 2006 | |
| Removal of zinc with EDTA | 3.03 | WT | highly destabilizing | Bullock AN, 2000 | |||
| T123A/H168R | 3.12 | WT | highly destabilizing | Nikolova PV, 2000 | |||
| T123A/H168R/R249S | 1.91 | WT | destabilizing | Nikolova PV, 2000 | |||
| T123A/M133L/H168R/V203A/ N239Y/R249S/N268D |
2.18 | T | destabilizing | Joerger AC, 2005 | 2BIQ | Joerger AC, 2005 | |
| T123A/R249S | 2.09 | WT | destabilizing | Nikolova PV, 2000 | |||
| M133L/H168R/V203A/N239Y/ R249S/N268D |
1.88 | T | destabilizing | Joerger AC, 2005 | 2BIP | Joerger AC, 2005 | |
| M133L/N239Y/N268D | -2.23 | WT | highly stabilizing | Nikolova PV, 1998 | |||
| M133L/V203A | -0.65 | WT | stabilizing | Nikolova PV, 1998 | |||
| M133L/V203A/N239Y/N268D | -2.65 | WT | highly stabilizing | Nikolova PV, 1998 | 1UOL | Joerger AC, 2004 | |
| V143A/N268D | 2.25 | WT | destabilizing | Nikolova PV, 2000 | |||
| H168R/R249S | 1.72 | WT | destabilizing | Nikolova PV, 2000 |
The H168R/R249S double mutation largely reverses the structural
perturbations caused by the single point mutations H168R and R249S. The guanidinium group of
Arg-168 in the
double mutant mimics the structural role of Arg-249 in the wild type that is key to
stabilizing the hairpin
conformation of the L3 loop required for sequence-specific DNA binding. As a result, the
wild type
conformation of the L2/L3 loop region is largely restored, and wild-type like DNA-binding
properties are
observed in the folded state (PDB 2BIP and 2BIQ).
|
||
| V203A/N239Y/N268D | -2.41 | WT | highly stabilizing | Nikolova PV, 1998 | |||
| Y236F/T253I | -1.6 | WT | stabilizing | Canadillas JM, 2006 | |||
| N239Y/G245S | 0 | WT | wild-type like | Nikolova PV, 2000 | |||
| N239Y/N268D | -2.04 | WT | highly stabilizing | Nikolova PV, 1998 |